 ClusterRows
ClusterRows
ClusterRows is a LibreOffice Calc extension that clusters the rows in a table and colors them to indicate the clusters.
Calc
Macro
Download latest
latest release: 2024-03-15 13:40:13
Description
How to use?
1. Click the toolbar item named Cluster rows which is next to the AutoFilter item or click on the 'Clustering...' menu item under Data > Statistics.
2. Now a dialog will appear where the input data cell-range(with or without header), the output location and the parameters of clustering can be set. If the input cell range was selected before launching the dialog then these two fields will be pre-filled. By default the output location is set to the column next to the last column of the input data cell-range for convenience. If the first row of the input range has the column headers then it can be specified by checking the 'Header in the first row' checkbox. It is also possible to specify whether the data rows need to be colored according to the cluster assignments.
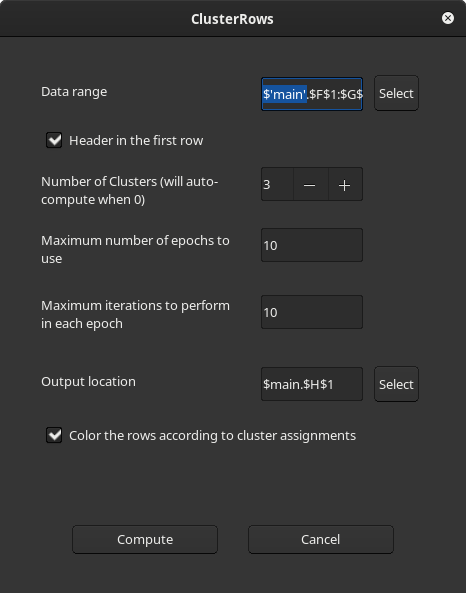
3. After pressing the *Compute* button, two new columns [ClusterId and Confidence] will be written to the user specified output location. These two columns will have headers if the dialog option `Header in the first row` was checked. The first column **ClusterId** specifies the cluster to which the row is assigned and the second column **Confidence** indicates the algorithm's confidence in scale [0,1] that this cluster assignment may be correct (higher number implies higher confidence). Depending on the choice provided in the dialog, the data rows are colored according to the cluster assignments.
Advanced usage via `GMMCLUSTER` formula
The dialog based clustering (described above) uses an in-house array formula `GMMCLUSTER` to compute the clusters and confidence scores. Hence it is possible to directly use this array formula to compute the clusters instead of using the dialog. Doing so has the advantage of specifying the exact data-range (without the header) and placement of the results. In addition the parameters (including the data-range) could be specified programatically as functions of other cells/ranges in general. The syntax of `GMMCLUSTER` is:
GMMCLUSTER(data, numClusters, numEpochs, numIterations)
where **data** is the array(cell-range) holding the data, **numClusters** is the desired number of clusters (optional, default is to automatically estimate this), **numEpochs** is the maximum number of epochs to use (optional) and **numIteration** is the maximum number of iterations to do in each epoch (optional). Note that after entering the formula expression remember to press `Ctrl+Shift+Enter` instead of just `Enter` to commit the array formula.
Implementation
The project uses an in-house C++ implementation of full Expectation Maximization algorithm to compute the clusters. In the auto mode (when number of clusters is specified as 0) it chooses the number of clusters parameter via Bayesian information criterion.
The project does not depend on any machine learning or linear algebra libraries. Full source code is made available under GPL3 license.
Planned features
* Optional 'seed' (5th) parameter for `GMMCLUSTER()` to make the results reproducible.
* Add support for other common clustering algorithms.
Sample usage of the extension
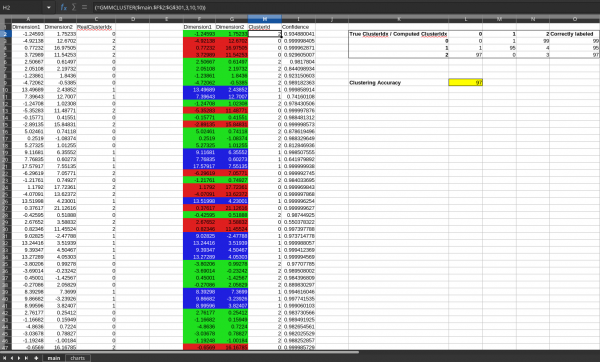
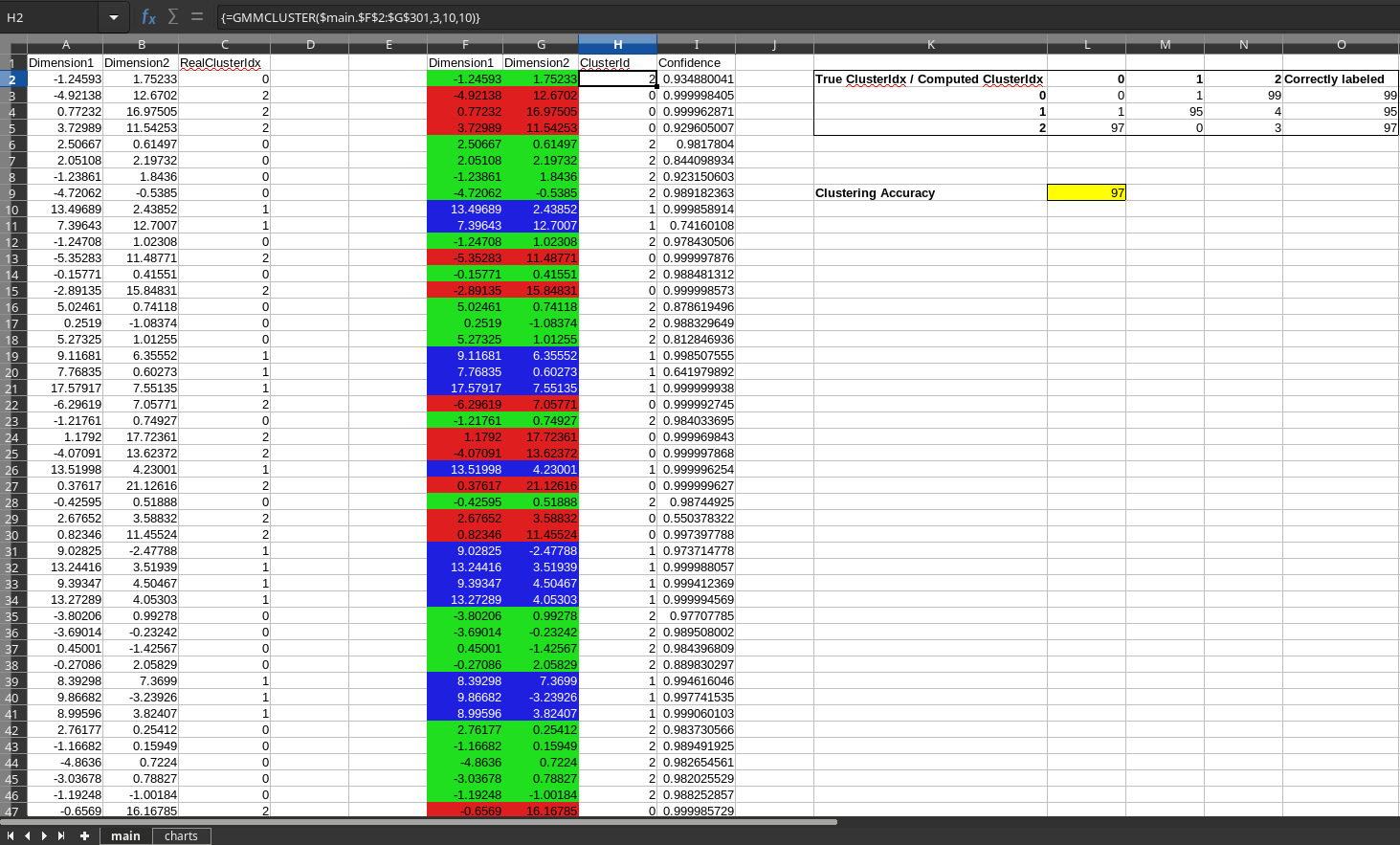
In 'testdocs' directory there is a spreadsheet file called 'three-clusters.ods'. In that sheet there is a dataset in the range `A1:C301` synthetically generated from a 3-cluster Gaussian mixture model. This toy dataset has two dimensions/variables(column A and B). Column C has the ground truth cluster id information of each row. To test the extension a copy of the dataset is placed(excluding the ground-truth column) at `F1:G301`. Go to the menu `Data > Statistics > Clustering...` and select or enter the input data range `F1:G301` and check the option `Header in the first row` and click `Compute` button in the dialog. The extension will compute the 3 clusters and produce the columns `ClusterId` and `Confidence` next to the data. The cell `L9` in that sheet will indicate the clustering accuracy. This is a measure of how well the clustering algorithm was able to assign clusters compared to the ground truths in the original dataset. Typically we get around 97% accuracy for this dataset.
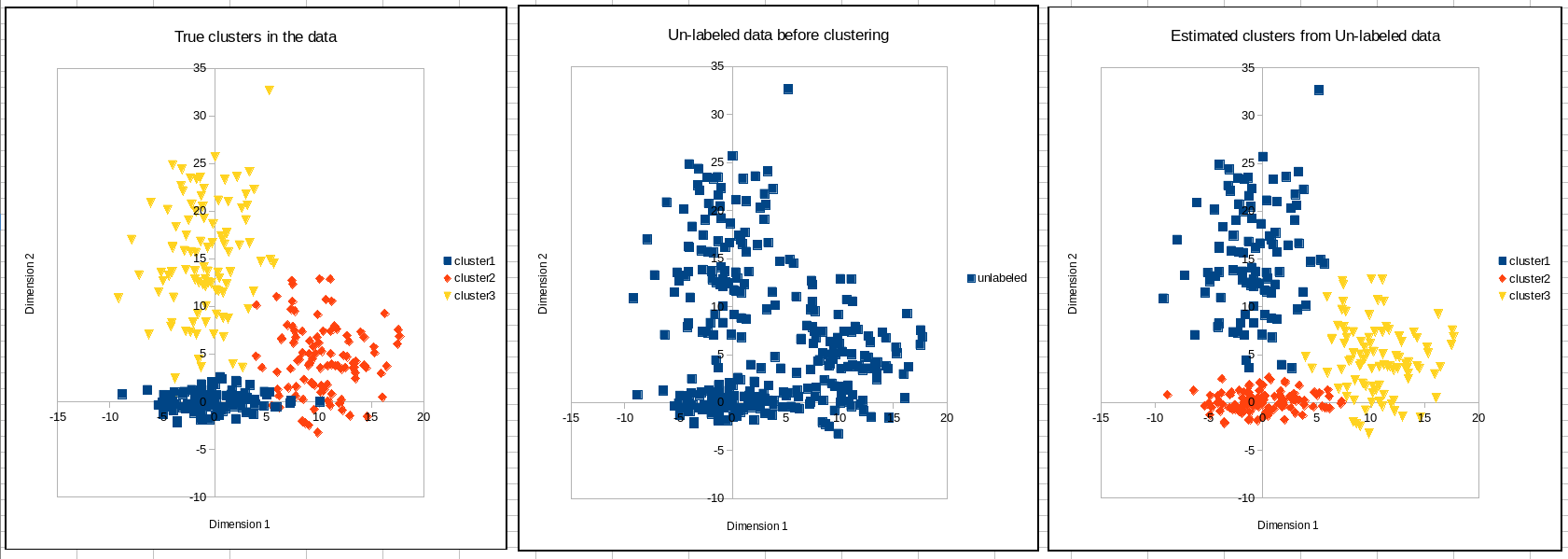
The document also contains visualizations of the data in the sheet `charts`. The left chart shows the data points colored according to the ground truth clusters of the dataset. The middle chart shows the data points without the cluster information (which is the input to the clustering algorithm). The right chart shows the data colored according to the cluster assignments made by the algorithm.
Homepage: https://github.com/dennisfrancis/ClusterRows
Repository: https://github.com/dennisfrancis/ClusterRows
Release List
Release
Description
Compatibility
Operating Systems
License
Release notes
2.0.0 Windows
Improvements and fixes
6.1
Windows
GPL
- Support for Windows and MacOSX
- Platform specific extension packages in the releases page.
- Dialog based workflow
- GMMCluster() formula
- Input range and the output location may be selected interactively and edited in the dialog
Download 2.0.0 MacOS Improvements and fixes 6.1 macOS GPL
- Support for Windows and MacOSX
- Platform specific extension packages in the releases page.
- Dialog based workflow
- GMMCluster() formula
- Input range and the output location may be selected interactively and edited in the dialog
Download 2.0.0 Linux Improvements and fixes 6.1 Linux GPL
- Support for Windows and MacOSX
- Platform specific extension packages in the releases page.
- Dialog based workflow
- GMMCluster() formula
- Input range and the output location may be selected interactively and edited in the dialog
Download
☆ ☆ ☆ ☆ ☆
Post your review
You cannot post reviews until you have logged in. Login Here.
Reviews
No one has commented on this page yet.
RSS feed for reviews on this page | RSS feed for all reviews